The usage tutorial
Two tools including SimDisExplore and SimPDExplore
are provided in DisSim. The details about the usage of these two tools are described as follows.
1. An example for using SimDisExplore.
SimDisExplore can be accessed from the web http://123.59.132.21:8080/DisSim/single.jsp. The following figure shows a case for searching similar diseases of ‘acute myocardial infraction’ and finding correlated TCs of ‘acute myocardial infraction’ and its similar diseases.
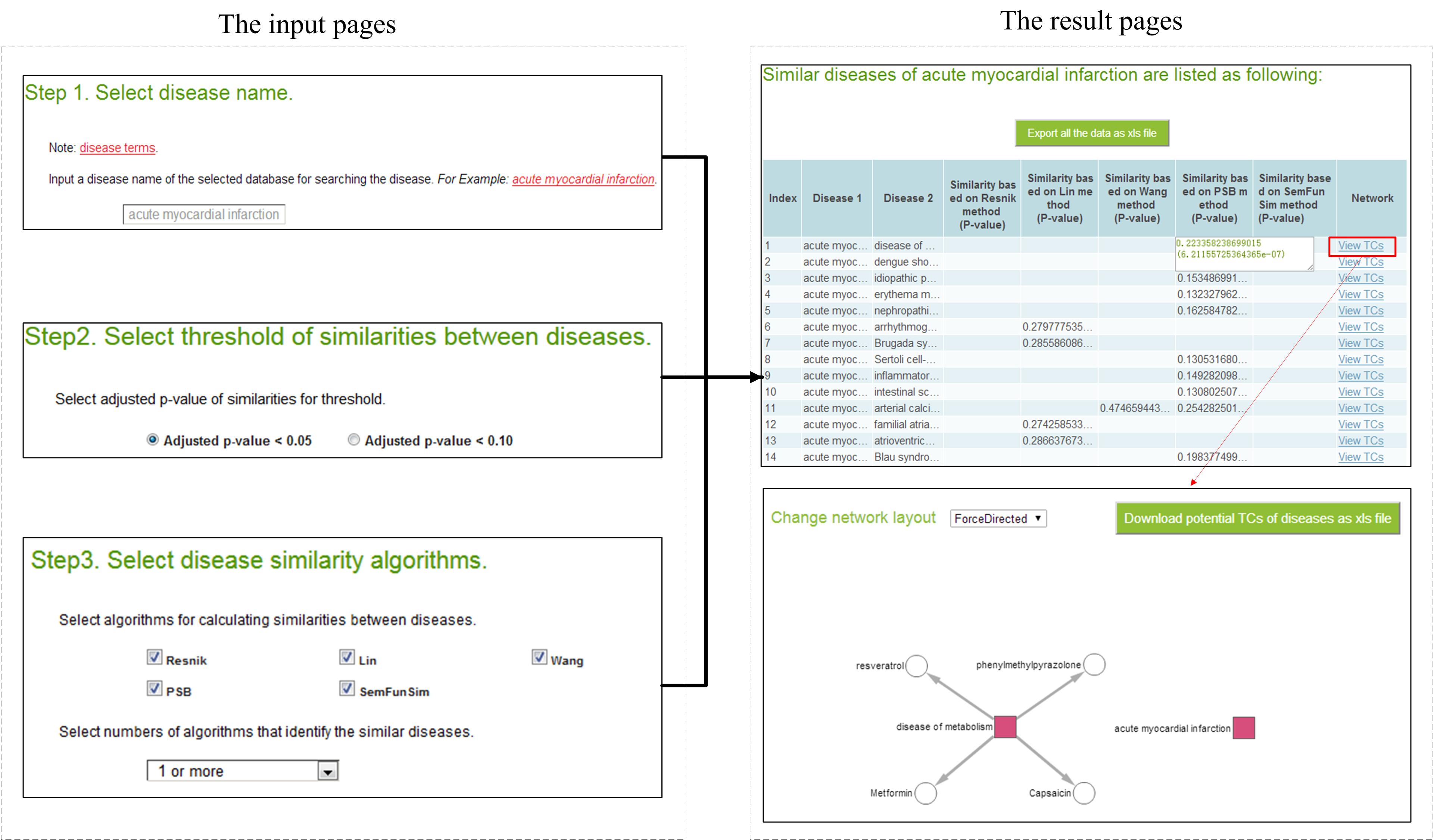
Searching similar diseases of 'acute myocardial infraction'.
The input page
http://123.59.132.21:8080/DisSim/single.jsp.
Step 1: Input a disease term. Disease terms of Disease Ontology (DO) in DisSim (see Materials and Methods) can be used as reference when inputting a disease term. These disease terms can be downloaded from the page. For convenience, we also provide the function to autocomplete the disease term.
In this case, we explore similar diseases of 'acute myocardial infraction'.
Step 2: Select a threshold for the similarity score. For an inputted disease term, the system will return its similar diseases with the P-value less than 0.10 or 0.05 according to the user’s selection. In this case, we choose the P-value less than 0.05 as the threshold.
Step 3: Select disease similarity algorithms. For an inputted disease term, five algorithms can be used to explore its similar diseases in DisSim. Users can select multiple algorithms as they need, and select the frequency of the pair that has been identified as similar diseases. In this case, all of these five algorithms are selected, and the frequency is selected as ‘1 or more’. It means that similar diseases identified by any one of these five algorithms would be shown.
After submitting the input page, similar diseases of the inputted disease term based on the selected algorithms are listed. The first column represents the number of similar diseases. The second and third columns represent the inputted disease and its similar diseases, respectively. The last column is the link to the network visualization of the relationships among TCs and the pair of diseases in this line. Each of the other columns lists the similarity score based on an algorithm and the P-value of the similarity score. All the results can be downloaded from the page.
2. An example for using SimPDExplore.
The following figure shows an example for searching similarity score and finding
correlated TCs between ‘liver disease’ and ‘gallbladder disease’.
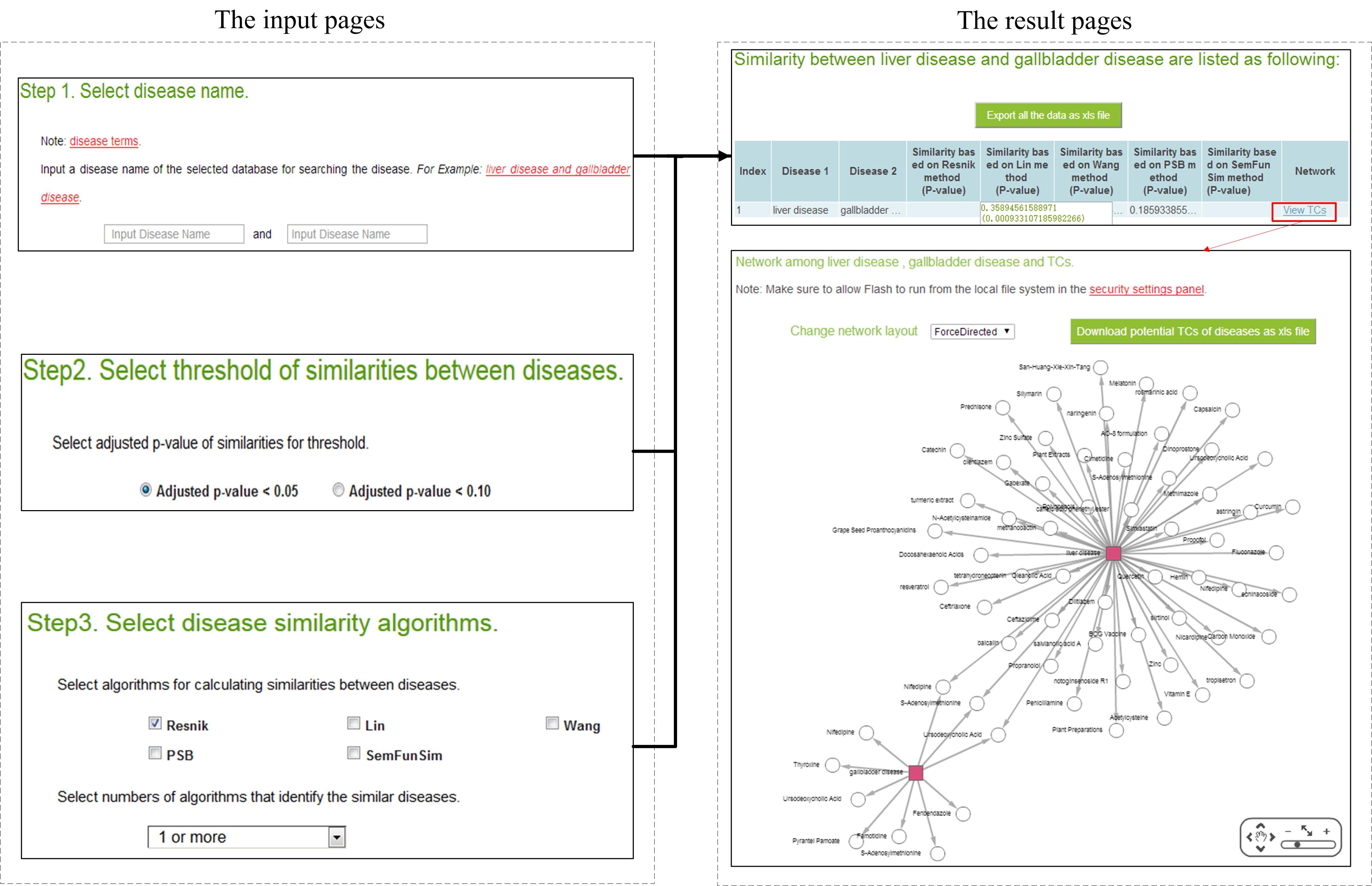
The input page of SimPDExplore can be accessed from the web
(http://123.59.132.21:8080/DisSim/pairs.jsp).
The difference is that the input page of SimPDExplore receives a pair of diseases.

