Introduction
Introduction of SV4GD
How to use SV4GD?
1. For browsing
Step 1:select one of disease category.
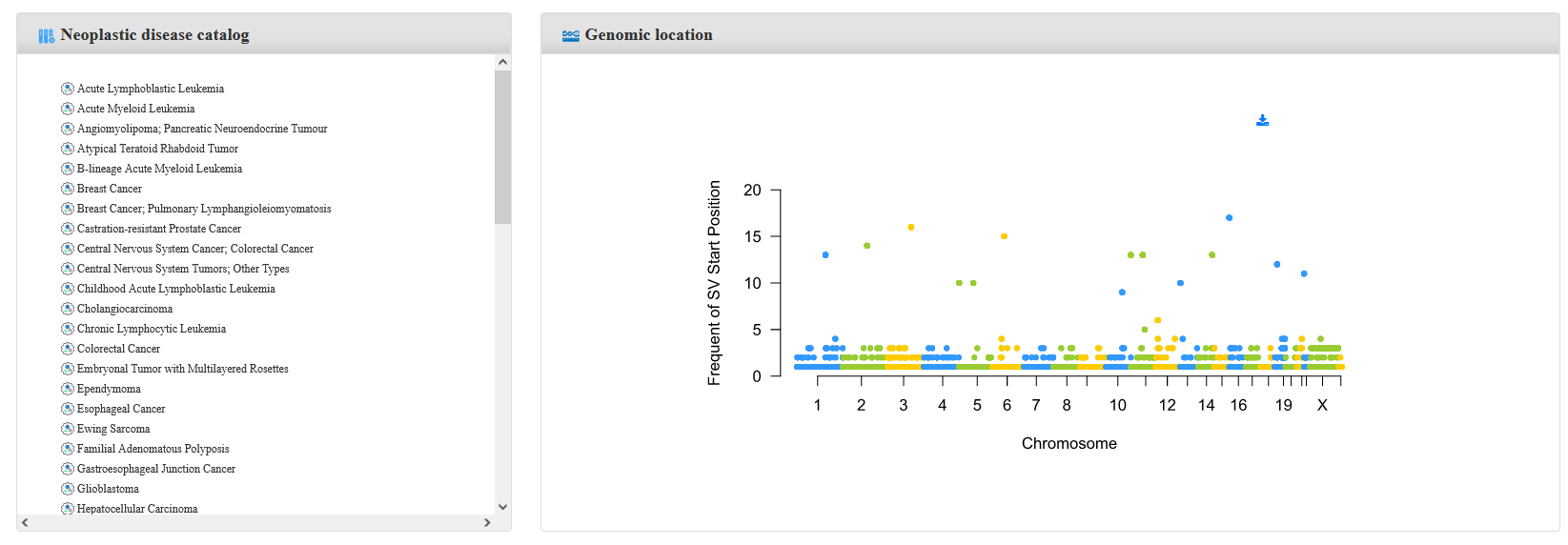
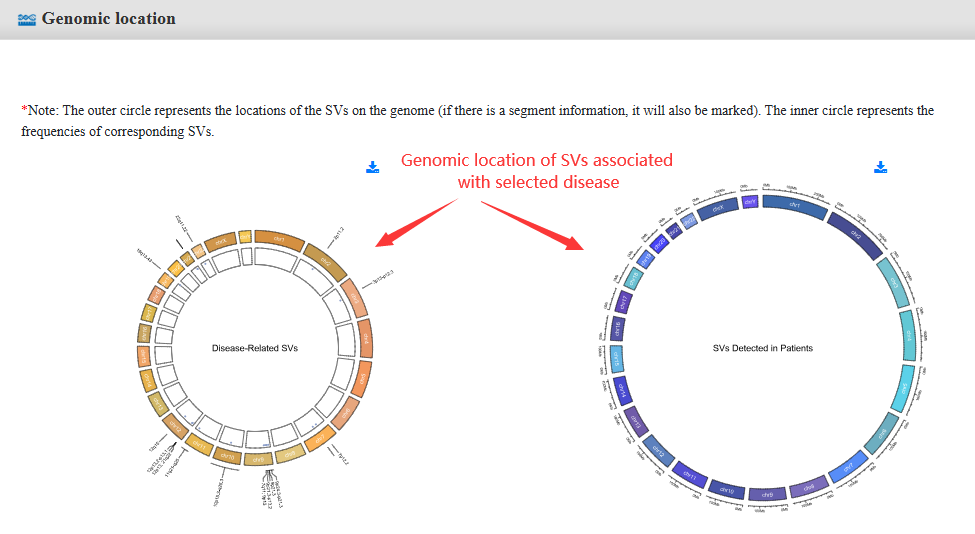
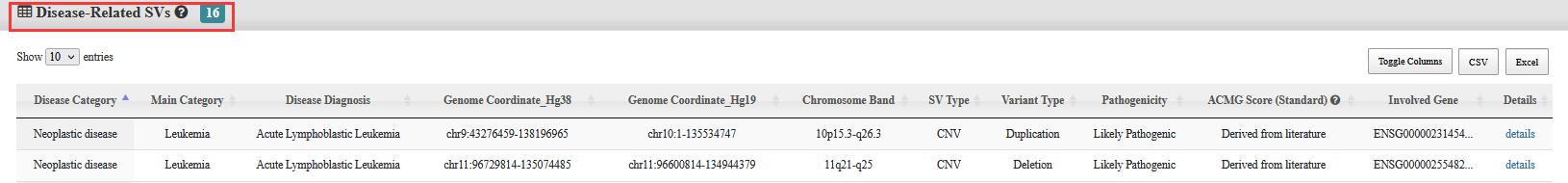
Step 2:Click on the interested specific disease, and the genomic distribution will display accompanied with two tables of the results, one is for Disease-Related SVs and one is for SVs Detected in Patients.
2. For searching
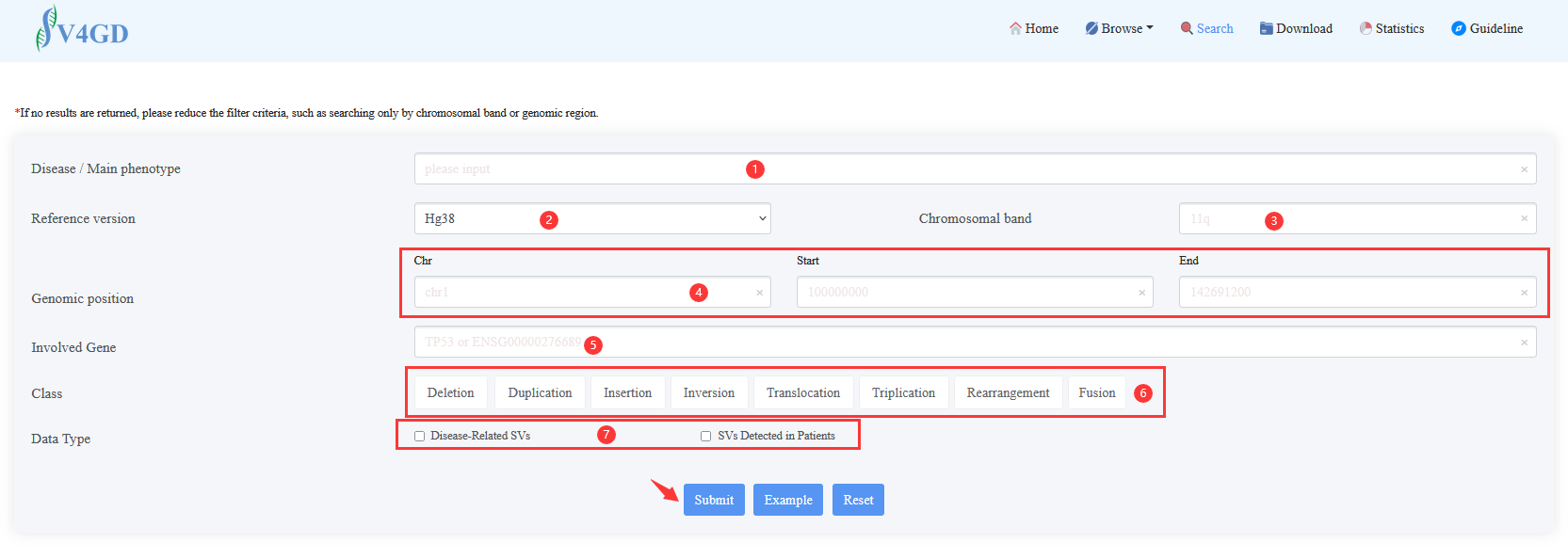
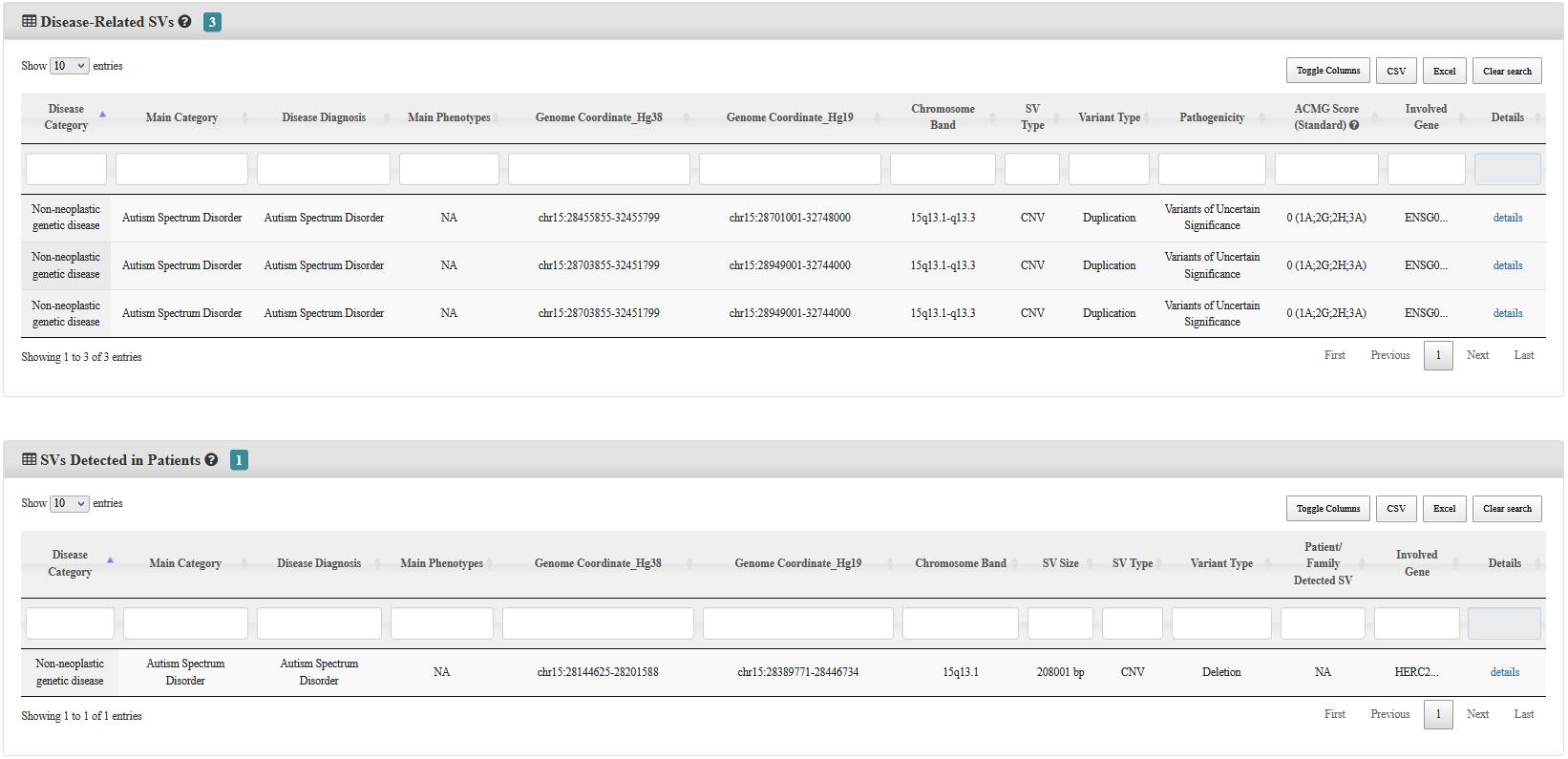
Step 1:Select or input any information of ① to ⑥, and then click Submit. Two result tables will be returned, one is for Disease-Related SVs and the other one is for SVs Detected in Patients.
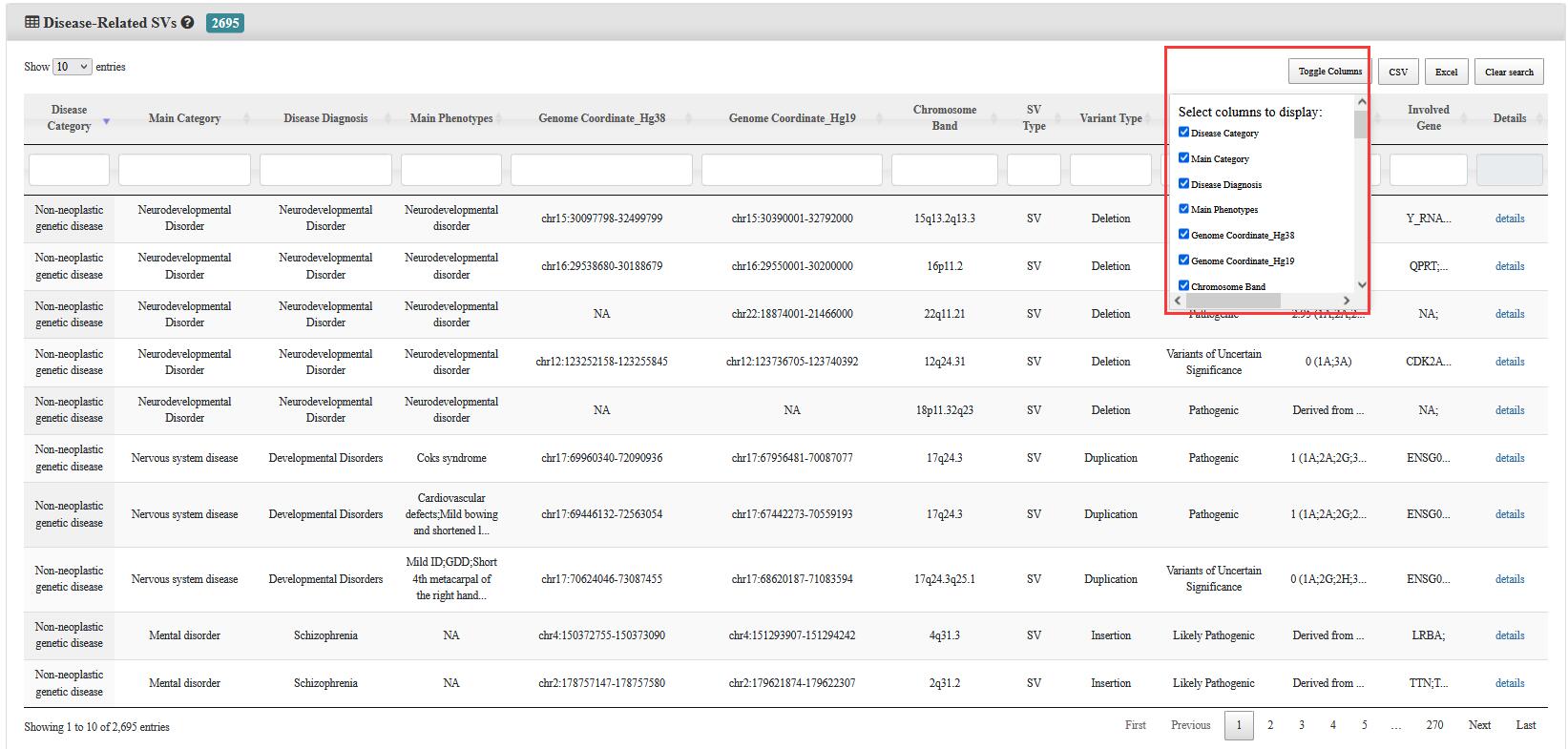
Step 2:Click on the Toggle Column, the user could select the interested information to be showed on the current page and download them in any form in the top right.
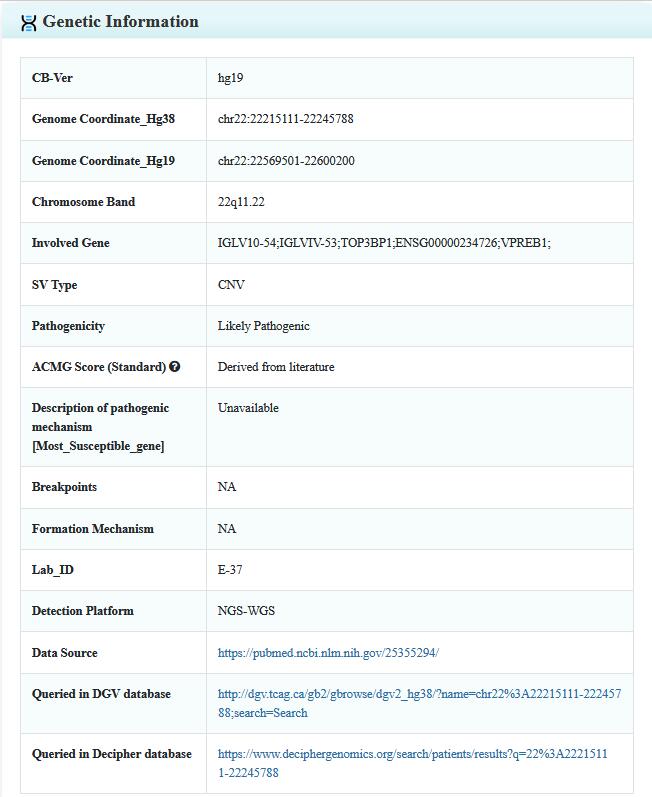
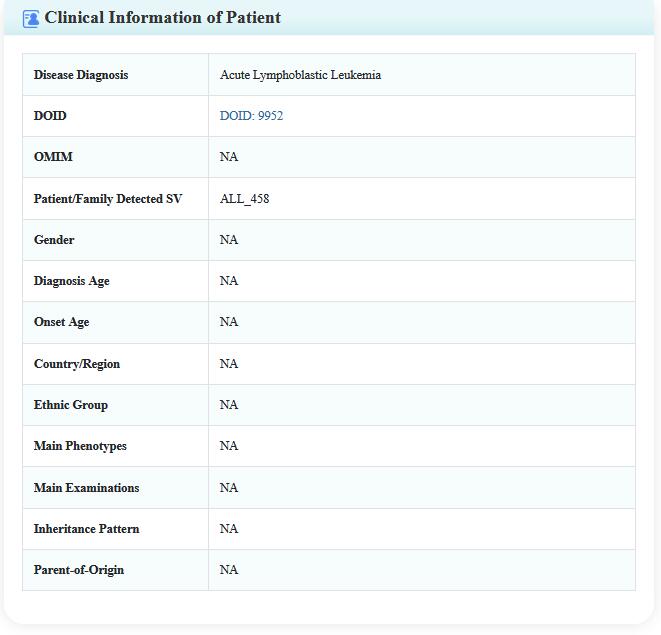
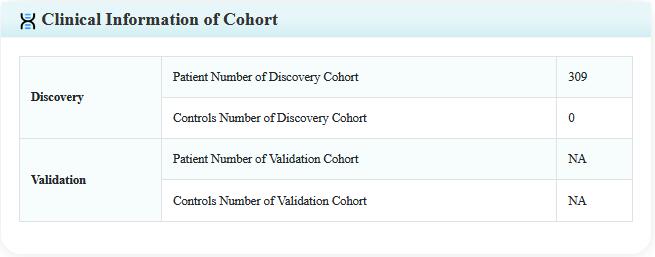
Step 3:Click on the detail button, the user will be leaded to a new page including the details of literature, variant, patient, and study cohort (for SVs Detected in Patients only).

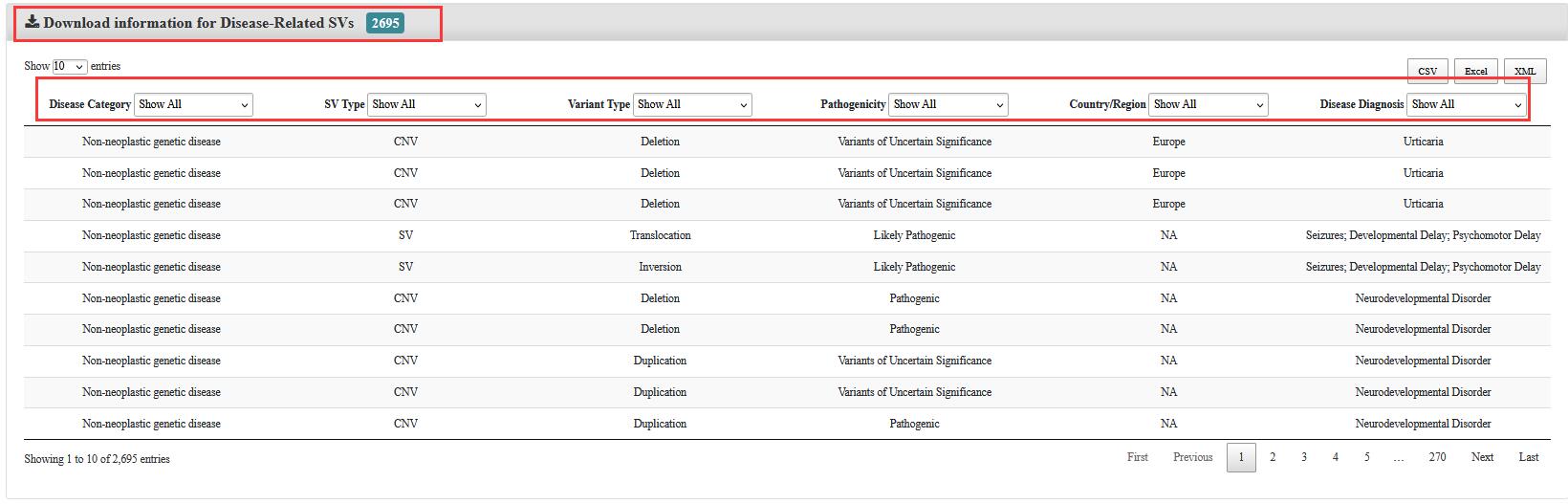
3. For downloading
The user could download the files in any format by clicking on the corresponding button.
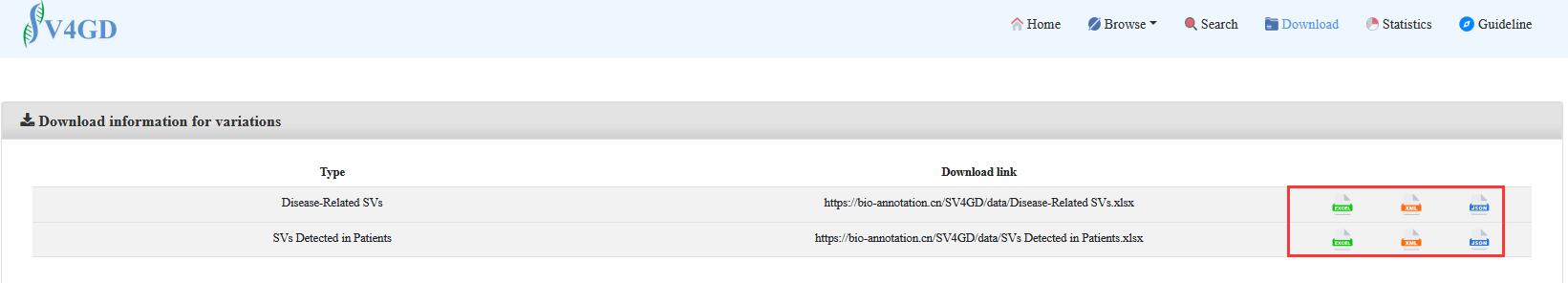
Meanwhile, if the user only wants to download the records interested in, the user could select the items step by step. and then obtain them through the button in the top right.
How to contribute to SV4GD?
And if the users found any errors or inconvenient usage while surfing on the SV4GD, please do not hesitate to contact us (Submit error correction prompts); we will appreciate your contribution to the improvement.
About us
We are a professional team aiming at the combination of basic experiments and bioinformatics dedicated to the analysis of bio-omics data, the development of algorithms, and the construction of biological databases. The databases currently developed by the laboratory are as follows:
gutMGene (https://bio-computing.hrbmu.edu.cn/gutmgene): a comprehensive database for target genes of gut microbes and microbial metabolites.
gutMDisorder (https://bio-computing.hrbmu.edu.cn/gutMDisorder): a comprehensive resource for associations between gut microbes and phenotypes or interventions in Human and Mouse, derived from manual literature extraction and raw data reprocessing.
Scovid (https://bio-computing.hrbmu.edu.cn/scovid): a comprehensive atlas for exposing the molecular characteristics of COVID-19 and other coronavirus.
microbioTA (https://bio-computing.hrbmu.edu.cn/microbiota): a microbial reannotation database that includes tissue microbial abundance data and comprehensive analysis results for human and mouse tumor tissue RNA sequencing data.
Details of our laboratory
Team
The SV4GD database was designed, created and maintained led by Lei Shi (Professor), Liang Cheng (Professor), and Xue Zhang (Professor, Academician of Chinese Academy of Engineering).
.png)

 Browse
Browse Search
Search Download
Download Statistics
Statistics Guideline
Guideline